 Researchers from the University of Washington have made use of online gamers to help decipher the structure of an AIDS-like retrovirus that scientists have been unable to break for fifteen years. The paper on the breakthrough in uncovering the crystal structure of M-PMV retroviral protease is published in the journal Nature Structural & Molecular Biology, with researchers and gamers both honoured as co-authors for their roles.
Researchers from the University of Washington have made use of online gamers to help decipher the structure of an AIDS-like retrovirus that scientists have been unable to break for fifteen years. The paper on the breakthrough in uncovering the crystal structure of M-PMV retroviral protease is published in the journal Nature Structural & Molecular Biology, with researchers and gamers both honoured as co-authors for their roles.
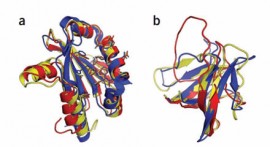
Previously scientists could only view the proteins as one-dimensional flat images, but pharmacologists need a three-dimensional view that “unfolds” the protein, giving them targets to potentially target with future drugs. FoldIt takes the problem of creating these 3D images, and outsources their creation to teams of online gamers who compete to unfold chains of amino acids to find the highest scoring (lowest energy) models.
It took the gamers just three weeks to refine the enzyme structure and researchers claim the discovery provides “new insights for the design of antiretroviral drugs”. It was a case of human intuition succeeding “where automated methods had failed” said Firas Khatib of the university’s biochemistry lab. Indeed, this method for discovery opens the door to making further use of online gamers, with the closing paragraph of the paper (embedded below) noting:
The critical role of Foldit players in the solution of the M-PMV PR structure shows the power of online games to channel human intuition and three-dimensional pattern-matching skills to solve challenging scientific problems…the ingenuity of game players is a formidable force that, if properly directed, can be used to solve a wide range of scientific problems.
Crystal structure of a monomeric retroviral protease solved by protein folding game players

